KAMILA Clustering on Big Mixed-Type Data
A significant proportion of studies investigating medical prescriptions have reported a rate of up to one-third for missingness of the physician’s specialty. In order to predict physician specialty, a study was designed utilizing our prescription database. Prior to conducting the prediction, it was necessary to explore and describe the data. The KAMILA clustering method was selected due to its ability to reduce heterogeneity and handle mixed-type data.
The data was imported into the R programming environment by establishing a connection between an SQL database and R using the DBI, ODBC, dbplyr, andtidyverse packages. Subsequent data wrangling and manipulation operations were performed. After the data was cleaned, there were a total of 17,137,949 prescriptions. Then, all combinations of two medications (2-combs) in every prescription were derived, and a new data set was developed.
Finally, KAMILA clustering was applied to the 2-combs data set. The forthcoming study will incorporate the utilization of supervised learning methodologies. Further details can be accessed here.
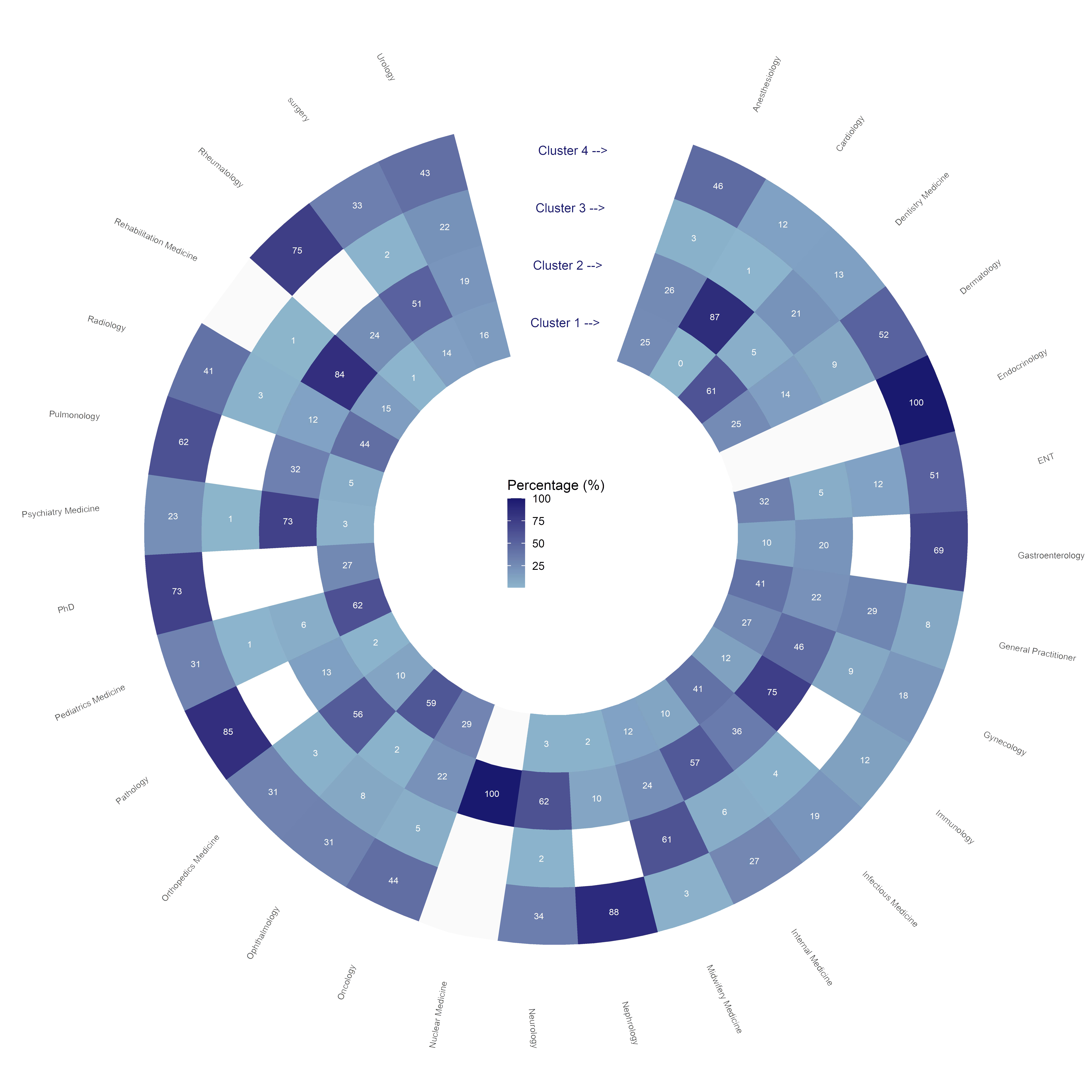
Figure 1 provides overall sight of the distribution of each specialty in four clusters. For example, endocrinology and nephrology were concentrated in cluster 1.

